- 124.25 KB
- 2022-08-12 发布
- 1、本文档由用户上传,淘文库整理发布,可阅读全部内容。
- 2、本文档内容版权归属内容提供方,所产生的收益全部归内容提供方所有。如果您对本文有版权争议,请立即联系网站客服。
- 3、本文档由用户上传,本站不保证质量和数量令人满意,可能有诸多瑕疵,付费之前,请仔细阅读内容确认后进行付费下载。
- 网站客服QQ:403074932
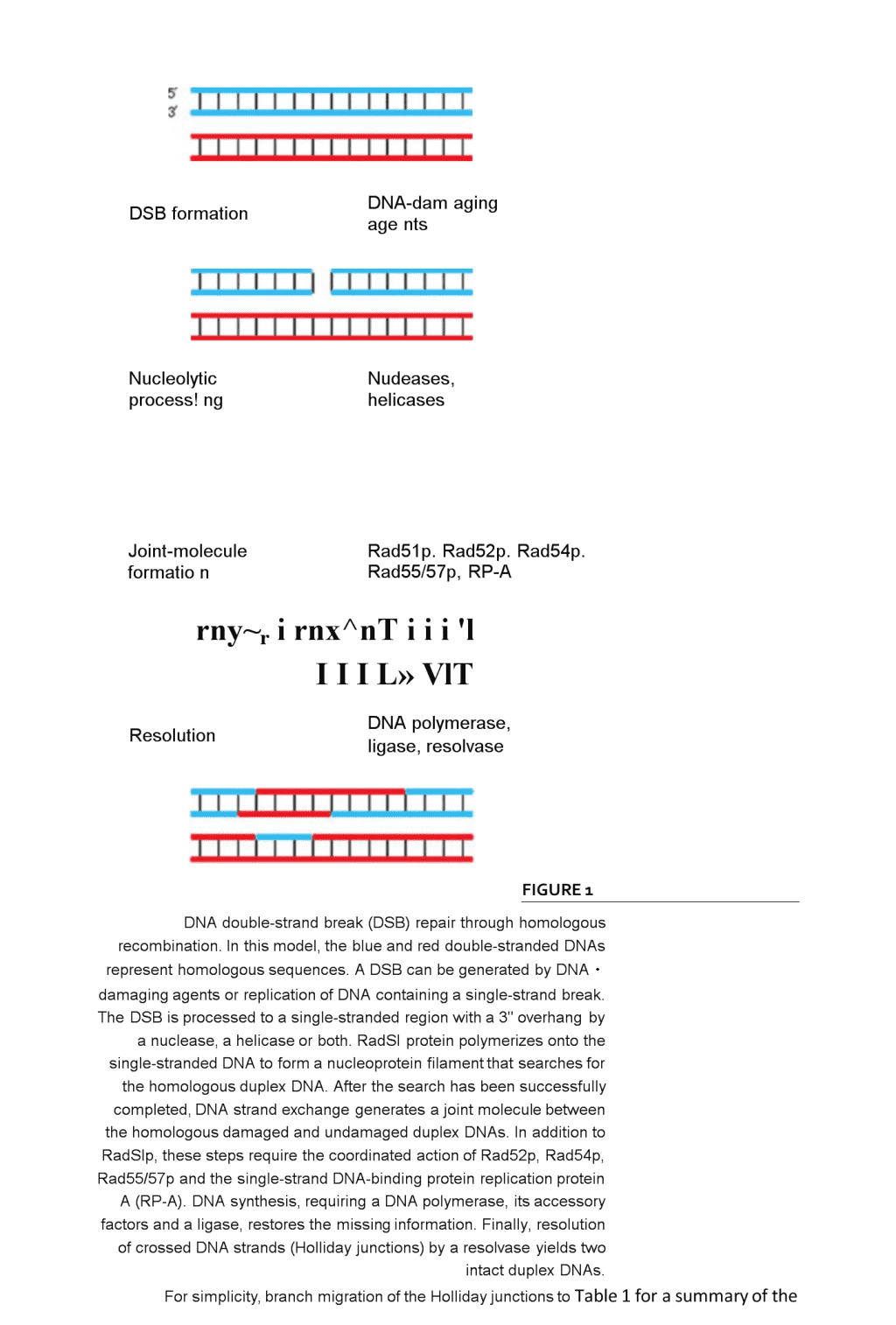
MolecularmechanismsofDNAdouble・strandbreakrepairRolandKanaar,JanH.J.HoeijmakersandDikC.vanGenDNAdouble・strandbreaks(DSBs)aremajorthreatstothegenomicintegrityofcells.Ifnottakencareofproperly,theycancausechromosomefragmentation,lossandtranslocation,possiblyresultingincarcinogenesis.UponDSBformation,cell-cyclecheckpointsaretriggeredandmultipleDSBrepairpathwayscanbeactivated.RecentresearchontheNijmegenbreakagesyndrome,whichpredisposespatientstocancer,suggestsadirectlinkbetweenactivationofcell-cyclecheckpointsandDSBrepair.Furthermore,thebiochemicalactivitiesofproteinsinvolvedinthetwomajorDSBrepairpathways,homologousrecombinationandDNAend-joining,arenowbeginningtoemerge.ThisreviewdiscussesthesenewfindingsandtheirimplicationsforthemechanismsofDSBrepair.TheintegrityofDNAinsidecellsisconstantlybeingchallengedbyendogenousandexogenousDNA-damagingagents.BecausealargevarietyoflesionscanoccurinDNA,itisnotsurprisingthatmultiplepathwayshaveevolvedthateachrepairasubsetoftheselesions1•ThesignificaneeofDNArepairisillustratedbythephenotypesofxerodermapigmentosum,Cockayne'ssyndrome,trichothiodystrophyandhereditarynonpolyposiscolorectalcancerpa2,3.ThesedisordersarecausedbymutationsinDNArepairgenesthatpredisposethepatientstocancer,neurologicalabnormalitiesorboth.Inaddi-tiontoefficientDNArepair,correctactivationofcell-cyclecheckpointsuponinductionofDNAdamageisofcrucialimportaneeforthemaintenanceofgenomicintegrity.CheckpointsallowactivelydividingcellstopauseandrepairDNAdamagebeforesegregationofthereplicatedgenomeintodaughtercells.TheirimportaneeisunderscoredbyinheriteddisordersassociatedwithdefectsinactivatingcellcyclecheckpointssuchasataxiatelangiectasiaandNijmegenbreakagesyndrome4,5.ThesedisorderscausehypersensitivitytoDNA-damagingagentsandspontaneouschromosomalinstability.Inthisreview,wefocusonmechanismsofDNAdouble-strandbreak(DSB)repairDSBsaregeneratedbyendogenouslyproducedradicalsandexogenousagentssuchasionizingradiation(IR),whichisoftenusedinanti-cancertherapy・RepairofDSBsisofcardinalimportaneetopreventchromosomalfragmentatio①translocationsand\ndeletions.Inthesoma,thegenomicinstabilityresultingfrompersistentorincorrectlyrepairedDSBscanleadtocarcinogenesisthroughactivationofoncogenes,inactivationoftumour-suppressorgenesorlossofheterozygosity,whileinthegermlinetheycanleadtoinborndefects.ThedeleteriouseffectsofDSBshavetriggeredtheevolutionofmultiplepathwaysfortheirrepair6,7.Homologousrecombinationrequiresextensivere・gionsofDNAhomologyandrepairsDSBsaccuratelyusinginformationontheundamagedsisterchromatidorhomologouschromosome・DNAend-joining,ontheotherhand,usesno,orextremelylimited,sequeneehomologytorejoinjuxtaposedendsinamannerthatneednotbeerrorfree(Figs1and2;see\nDSBformationDNA-damagingagentsNucleolyticprocess!ngNudeases,helicasesJoint-moleculeformationRad51p.Rad52p.Rad54p.Rad55/57p,RP-Arny~rirnx^nTiii'lIIIL»VlTResolutionDNApolymerase,ligase,resolvaseFIGURE1DNAdouble-strandbreak(DSB)repairthroughhomologousrecombination.Inthismodel,theblueandreddouble-strandedDNAsrepresenthomologoussequences.ADSBcanbegeneratedbyDNA・damagingagentsorreplicationofDNAcontainingasingle-strandbreak.TheDSBisprocessedtoasingle-strandedregionwitha3"overhangbyanuclease,ahelicaseorboth.RadSIproteinpolymerizesontothesingle-strandedDNAtoformanucleoproteinfilamentthatsearchesforthehomologousduplexDNA.Afterthesearchhasbeensuccessfullycompleted,DNAstrandexchangegeneratesajointmoleculebetweenthehomologousdamagedandundamagedduplexDNAs.InadditiontoRadSlp,thesestepsrequirethecoordinatedactionofRad52p,Rad54p,Rad55/57pandthesingle-strandDNA-bindingproteinreplicationproteinA(RP-A).DNAsynthesis,requiringaDNApolymerase,itsaccessoryfactorsandaligase,restoresthemissinginformation.Finally,resolutionofcrossedDNAstrands(Hollidayjunctions)byaresolvaseyieldstwointactduplexDNAs.Forsimplicity,branchmigrationoftheHollidayjunctionstoTable1forasummaryofthe\npropertiesoftheproteinsinvolvedandthecorrespondingmutantphenotypes).GeneticanalysesoftheeffectsofIRonSaccharomycescerevisiaeandrodentcellshavebeencrucialinidentifyinggenesinvolvedinDSByeastmutantsweredefectiveinhomologousrecombination,whereasthemutantrodentcelllinesweredefectiveinDNAend-joining.However,recentstudieshaveshownthatbothpathwayscontributetoDSBrepairinyeastandmammalsbutthattheirrelativecontributioncanvary(seebelow).ThegeneticanalysesarenowbeingcomplementedbythebiochemicalcharacterizationoftheDSBrepairpathways・DSBrepairbyhomologousrecombinationGeneticanalysesinS.cerevisiaehaveledtotheidentificationofgenesrequiredforDSBrepairbyhomologousrecombination9,12.Screensforradiation・sensitiveormeioticrecombination-deficientyeastcellshaverevealedtheinvolvementoftheRAD52epistasisgroupgenes(RAD50,RAD51,RAD52,RAD54,RAD55,RAD57,RAD59,MREllandXRS2).TheimportanceoftheRAD52recombi・nationalDNArepairpathwayisunderscoredbyitsevolutionaryconservation.MouseandhumangeneswithsequencesimilaritytoRAD50,RAD51,RAD52,RAD54andMREllhavebeenisolated7,10・Theavail-abilityoftheRAD52groupgeneshasprovidedtheessentialtoolsforbiochemicalanalysesofthemolecularmechanismofhomologousrecombination.TheaminoacidsequenceofS.cerevisiaeRad51pguidedtheinitialbiochemicalexperimentsbecauseofitssimilaritytotheEscherichiacolirecombinationproteinRecA6•Theseexperimentsshowedthatkeyreactionsinhomologousrecombination一thesearchforhomologousDNAandDNAstrandexchangeareperformedbyremarkablysimilarmechanismsinbacteria,yeastandhumancells.RecA,Rad51pandhumanRAD51polymerizeonDNAtoformanucleoproteinfilamentthatsearchesforhomologousDNA13•ThissearchendswiththeformationofjointmoleculesbetweenthehomologousDNAs,followedbyexchangeofDNAstrands(seeFig.1).TheS.cerevisiaeproteinsRad55pandRad57pdisplaysequeneesimilaritytoRad51p・However;itisunlikelythatthesearefunctionalhomologuesofRad51psincethereisnoevidenceforRad55porRad57pnucleoproteinfilamentformation.Instead,thesetwoproteinsformaheterodimerthatcanstimulateRad51p-mediatedstrandexchangeatsub-stoichiometricamountsrelativetoRad51p14.ThereareseveralotherproteinswithsimilaritytoRadSlpinmammalsaswell.SincedisruptionofmouseRAD51resultsinembryoniclethality6,theothermammalianproteinswithsequencesimilaritytoRAD51(RAD51B,RAD51C,RAD51D,XRCC2andlossofmouseRAD51functionXRCC3)areapparentlyunabletosubstituteforthe15-21•Atpresent,itisunclearwhethertheseproteinscanformnucleopro-einfilaments,likeRAD51,orwhethertheyfunctionanalogouslytoRad55/57p.ThereareseveralreasonswhymultipleRAD51-likeproteinsthatcanformnucleoproteinfilamentsmightberequiredinmammals.•Theycouldassembleseparatenucleoproteinfila-mentsthathavespec讦iccharacteristicstofacilitaterepairinpartsofthegenomethatdifferinchromatinstructureorcontainspecialsequencessuchastelomeresorcentromeres.Inaddition,nucleoproteinfilamentswithslightlydifferentpropertiesmightberequiredtorepairdifferenttypesofDNAdamage,suchasDSBsorinterstrandDNAcrosslinks.•Theycouldformmixedfilamentsinwhichthedifferentproteinshavespecificfunctio\nns.Forexample,aRAD51-likeprotomercouldbeincor・poratedintheRAD51nucleoproteinfilamentatsubstoichiometriclevelstoserveasadockingsiteforotherproteinsthatneedtointeractwiththefilament,butnotwitheveryRAD51protomer.•Theycouldberequiredwhenthehomologouschromosomeinsteadofthesisterchromatidisusedasatemplate.•Theproteinscouldhavetissue-specificfunctions.ThestepsthatleadtoS.cerevisiaeRad51p-mediatedjoint-moleculeformationcanbestimulatedbyovercomeinhibitoryeffectsofcompetingreactionsinthecellInadditiontoRad55/57p,Rad52pplaysacrucialroleinstimulatingRadSlp.BiochemicalanalysesofRad52phaveshownthatitpromotesannealingofcomplementarysingle-strandedDNAs22andfunctionsasamediatorbetweenRadSlpandthesingle-strandDNA-bindingproteinRP-Atostimu-lateRad51p-mediatedDNAstrandexchange14,23,24Similarly,humanRAD52proteinassistshumanRAD51informingjointmolecules25•However,thephenotypeofRAD52mutationsinS.cerevisiae,fissionyeastSchizosaccharomycespombeandmousecellsdiffersdramatically.WhiletheefficiencyofrecombinationisreducedbymorethanthreeordersofmagnitudeintheS.cerevisiaerad52mutant,itisonlytwo・foldlowerinthecorrespondingS・pombemutantandinmouseRAD52-knockoutembryonicstemcells7,26.TheproteinencodedbytheS.cerevisiaeRAD59geneshowssequeneesimilaritytoRad52p27.AdditionalhomologuesofRAD52andRAD59havenotyetbeenidentifiedinotherspecies,buttheexisteneeofRAD59homologuesinotherspeciescouldhelpexplainthediffereneeinphenotypesconferredbyRAD52ifsomeofthefunctionsofRAD52canalsobeassumedbyRAD59inS.Pombeandmammals-Anotherkeycomponentofthehomologous-recombinationmachinery,theDNA-dependentATPaseRad54p28,29,belongstotheSNF2-SWI2proteinfamily6.MembersofthisfamilyareinvolvedinmanyastsofDNAmetabolism,suchastranscription,recombinationandDNArepair.GeneticexperimentshaveshownthatRAD54-knockoutyeast,chickenandmousecellsareIR-sensitiveandhaveareducedlevelofhomologousrecombination12,30,31.Rad54pcaninteractwithRad51p32-34,andbiochemicalexperi・meritshaveshownthatRad54pstimulatesRad51pmediatedjoint-moleculeformation28.BecauseRad51pactivitycanbestimulatedbyanumberofproteins(Rad52p,Rad54pandRad55/57p),itisofinteresttodeterminetheexactstepinthestrand-exchangereactionatwhichtheseproteinsact.TheotherproteinsintheRAD52epistasisgroup,Rad50p,MrellpandXrs2p,formacomplexthatisrequiredfortheformationofmeiosis-specifiDSBs,whicharesubsequentlyrepairedthroughrecombinationwiththehomologouschromosome9・Inmitoticcells,thecomplexisprobablyinvolvedinonlyasubsetofDNArepairreactionsthatrequirehomologousrecombination35.Interestingly,recentexperimentshaveshownthatRad50p,MrellpandXrs2parealsorequiredforDNAend-joining(seebelow).DSBrepairbyDNAend-joiningTheDNAend-joiningpathwayofDSBrepairwasfirstcharacterizedinrodentcells36•UsingacollectionofChinesehamsterovarycelllinesselectedonhebasisoftheirradiosensitivity,threeDSBrepairgeneshavebeenisolated:XRCC4,XRCC5and\noODNA-PKCSSir2p/3p/4pRAD50MRE11NBS1LigaseIVXRCC4FIGURE2ModelforDNAdouble-strandbreak(DSB)repairthroughDNAend-joining.UponDSBformation,theKUheterodimerbindstotheDNAendsandattractsDNA-PK^.Inadditiontothis,chromatinstructuremightalsobeinfluenced.InSaccharomycescerevisiae,Sir2p,Sir3pandSir4pareinvolvedinthisprocess,andasimilarchromatin-remodellingreactionmightalsooccurinmammals.Subsequently,theendsarebroughttogetherandtheDNA-PKC$protomersphosphorylateeachother,causingastructuralchangeinthecomplex,possiblyresultinginremovalofDNA-PKCS.Forthelaterstagesoftherepairprocess,thecomplexcontainingRAD50,MRE11andNBS1isattracted,whichmightprocesstheends.Finally,theDNAligase-IV-XRCC4complexrejoinsthestrands.ItshouldbestressedthatitisnotknowninwhichorderthevariousproteincomplexesareattractedtotheDNAend.ThetwointerwoundDNAstrandsarerepresentedasribbons,andproteinsasspheres.Higher-orderchromatinstructureisnotindicated,althoughtheSirproteinsactbymodulatingchromatinstructurethroughinteractionswithhistones.\nreviewsTABLE1-DSBREPAIRPROTEINSANDMUTANTSOFSACCHAHOMYCtSCtRtVISIAtANDMAMMALS*SourceMutantphenotypesInteractionRemarks$・cerev/s/aiMammalianJ.cerev/s/aeMammalianRadSIpRADS1VeryIR-sensttivt11EmbryonklethaPx心<152护";RadS4pM-M#BRCA1SBRCA24SearchforDNAhomology*RadS2pRADS2VeryIR-sen$itive,2Slightlyreducedrecombination2*RadSIpStimulatesRJdS1p*A6RadS4pRAD54VeryIR-$ensitiveuIR-sensitiveEScelbwithreducedrxomb*u計on10川RadSIpStimulateRad51paRadS5/57pIR-sensithre12Rad51pStimulatesRadSIp14RADS1B-DSimilaritytoRADSI11-1*XRCC2andXRCC3DNAxDNA-endbinding'1"DNAI.8.492-494;gY.etol.(1997)Immunity7.6S3-66S;C.C.ftal.(1998)Mo/.CW/2,1-8;^HartJey,K.O.rfol.(199$)CeHB2,849-856;^Peterson,S.R.etal.(1995)P/oc.Natl.Mod.$dU.S.A.92.5171-3174;rBluntT.etM(199S)Ce«80.813-823;'CottNeb,T.M.andJackson,S.P.(1991)OV7X1B1-142;Herrmann,C・,Undahl,T.indSchir,P.(1998)£MflOA17,4188-4198;^Crawunder,U.etol.(1997)NW388,492-495;**Critchlow,S.E.#Bowater,R・P.andJackson,S.P.(1997)Curr.Bicl.7,588-598;neo,S.H.andJackson,S・P.(1997)fMBO/.16#4788-4795.\nXRCC7.XRCC5andXRCC7encodesubunitsofDNA-dependentproteinkinase(DNA-PK).Thethirdsubunitofthiscomplex,KU70,wasidentifiedthroughbiochemicalstudies(seebelow).DNA-PKisnotonlyrequiredforrepairofIR-inducedDNAdamagebutalsoforV(D)Jandimmunoglobulinclass-switchrecombinationintheimmunesystem37-41.MiceinwhichcomponentsofDNA-PKhavebeeninactivatedshowseverecombinedimmunodeficiency(SCID)aswellasIRsensitivity.BiochemicalanalysesoffactorsinvolvedinDNAend-joininghaveprovidedadditionalinsightintothemechanismofthisrepairreaction36.AheterodimerofKU80,encodedbytheXRCC5gene,andKU70bindswithhighaffinitytoDNAends,sug-gestinganearlyKUbindingprobablyroleinprotectsdamagerecognition,processingorboth,theendsfromnucleolyticdegradation42.Inaddition,KUproteinmightplayaroleinjuxtapositioningoftwoDNAendssinceitcantransferbetweenDNAfragmentswithcomplementaryoverhangs43andstimulatemammalianDNAligases44.Moreover,theshapeofafractionofKU-DNAcomplexesimagedbyscanningforcemicroscopyisconsistentwithanarrayoftwoDNAmoleculesheldtogetherbyKU45-47.UponbindingtoaDNAend,KUcanattractthecatalyticsubunit(DNA-PKCS),a465kDapolypeptidewithaC-terminalproteinkinasedo-main.AlthoughDNA-PKCShassomeDNA-dependentproteinkinaseactivityitself,theDNA-boundKUheterodimergreatlyenhancesthisactivity47,48.DNA-PKphosphorylatesmanydifferentproteinsinvitro,butitsinvivotargetsandthefunctionalsignificaneeofthephosphorylationsarestillunclear38AninterestingDNA-PKactivityisautophosphorylationofthecatalyticsubunit,whichresultsininactivationofitskinaseactivity,probablycausedbyreducedaffinityofphosphorylatedDNA-PKCSfortheKUheterodimer49・AmodelforaDNA-PKfunctionearlyinDSBrepairhasbeendevised(Fig.2).UponbindingtoaDNAenctKUattractsthecatalyticsubunit.Subsequently,theendsarebroughttogether,andtheDNA-PKmoleculeononeDNAendphosphorylatesthecatalyticsubunitlocatedontheotherDNAend.Thistransphosphorylationinactivatesthekinaseactivity,therebyabrogatingpossiblesignallingfunctions.Inaddition,thephosphorylationcouldresultinremodellingoftheproteincomplexontheendstoallowaccessofotherrepairfactorstothebreak.AdditionalactivitiesrequiredforDNAend-joiningineludeprocessingofDNAendsandligation.ProcessingofDNAendsiscurrentlyapoorlyunderstoodreaction.ItmightrequireanucleaseandaDNApolymerase,butnocandidategeneshavebeenidentifiedinmammals.Recently,DNAligaseIVhasbeenimplicatedinDNAend-joiningbecauseofitstightinteractionwithXRCC411•However,itcannotbeexcludedthatotherligasescould(partially)takeoveritsfunction.FurthergeneticanalysisoftheDNAend-joiningpathwayhasbeenfacilitatedbythediscoverythatitalsoexistsinS.Cerevisiae・HomologuesofKU70,KU80,XRCC4andDNAligaseIVhavebeenidentifiedandshowntofunctioninDNAend-joining.DeletionoftheseyeastgenesdoesnotcausehypersensitivitytoIR.However,ifhomologousrecombinationisinhibited,forexamplebydeletionofRAD52orinactivationoftherepairtemplate50,aneffectofScKU70orScKU80disruptioncanbeobserved.GeneticanalyseshaverevealedthatthesegenesareepistaticwithRAD50,MRE11andXRS251,52.Deletionmutantsofthesegenesjoincomplementaryendswithseverelyreducedefficiency.Biochemicalexperimentshave\nshownthathumanMRE11possesses3toexonucleaseactivity,suggest!ngthatthecomplexmightbeinvolvedinnucleolyticprocessingoftheendsbeforesubsequentrepair53.MammalianhomologuesofRad50pandMrellphavebeenidentified10.Theproteinsformacomplex,suggestingconservationoftheirrepairfunc-tions.AfterIR,theproteincomplexcontainingRAD50andMRE11concentratesinfociinthenu cleus(Fig.3)54•Uponpartialirradiationofthenucleusofhumanfibroblasts,thesefocioccurintheirradiatedvolumeonly,consistentwithadirectroleinDSBrepair55.ChromatinstructurecouldposeanadditionallevelofcomplexitytotheDNAend-joiningprocess.DeletionoftheSIR2,SIR3orSIR4genesinS.cerevisiaenotonlycausestranscriptionalderepressionoftelomericgenesandtelomereshorteningbutalsoleadstoinhibitionofDSBrejoining52,56•TheSirproteinsarerequiredformaintenanceofaspecificchromatinstructurethatrepressestranscriptionattelomeres.ItisconceivablethatasimilarchromatinstructurecouldfacilitateDNAend-joining.UnlikeDSBrepairbyhomologousrecombination,forwhichchromatinstructuremightcomplicatethesearchforhomology,repairbyDNAend-joiningcouldtakeadvantageofchromatinstructuretokeepthetwoDNAendsofaDSBincloseproximity.AhumanhomologueoftheSIR2genehasbeenidentified,indicatingthatsimilarfunctionsforchromatinstructuremightexistinmammaliancellsaswell11.Linksbetweencell-cycleregulationandDSBrepairAfterDSBformation,thecellcycleisarrested,followedbyrepair.AtaxiatelangiectasiaandNijmegenbreakagesyndrome,twocongenitalhumandisorders,arecausedbymutationsingenesinvolvedinthiscell-cyclearrest4,5.CellsfromthesepatientsarehypersensitivetoIRandfailtosuppressDNAsynthesisinresponsetoirradiation.OneofthemostexcitingrecentdevelopmentsinDSBrepairresearchistheidentificationoftheNBS1genethatismutatedinNijmegenbreakagesyndromepatients57-59.NBS1proteinwasfoundtobeoneofthecomponentsoftheRad50-Mrellcomplex,therebyphysicallylinkingDSBrepairproteinswithapotentialcell-cycleregulator,DNAdamagedetectionfactororboth(seeFig.3).ThiscomplexmighthaveacentraIroleinDSBrepair;inadditiontoaffectingthecellcyclethroughNBS1activity,thenucleaseactivityoftheMrellproteincouldfacilitateDNAendprocessing・\nreviews*/*...•.••ip■、*■NBS1■••*■・7MRE11DARI和;•MergedFIGURE>Double-strandDNAbreak-inducingagentscausetheformationofnucleartodcontainingNRS1andMRE11.NuclearfodofthehumanNBS1(p9SXMRE11andRADSOproteinsareinducedbytreatmentoffibroblastswrthiomangradutior^butarenotseenmuntreatedcellsst.ThefociarerestrictedtotheirradiMedvolumeofthenucleus11.TheNBS1andMRE11pioteinsxestainedgrten□ndr«4respectivety.ThenuckusisstainedwrthDAPt.ColoaltzationofNBS1ancMRE11isevidentfromtheye#owfociinthemergedimage.ThismuftlprotemcomplexlinksDNArepairwiththecellcycle.Thenuc^aseactivityofMRE11suggeststhxitmightbeinvolvedinnudtolyticproceisingofDNAendsbeforesubsequentrepjr11.CelhderivedfromNijmegenbrejfcjgesyndromepatientsthatcarrymutationsintheNtS1genezrehypersensitivetoionizingradMionjnddispbyspontaneouschromosomalinstability・TheyfailtosuppressDNAsyrthesisinresponsetoirradiatx)nowingtoidefectinactivatingacelkcyclecheckpoint5.SpecializedandadditionalfunctionsofDSBrepairpathwaysItisnotimmediatelyapparentwhymultiplepathwayshaveevolvedtorepairDSBs.OnereasoncouldbeprovidedbythepotentialdifferenceinfidelityofDSBrepairbyDNAend-joiningandhomologousrecombination.DNAend-joining,asimpleanddirectsolution,mightresultinerrorsasnucleotidesatthebreakcouldbeaddedorlostandincorrectendsmightbejoined.Infact,theimmunesystemexploitsthisinaccuracytocreateadditionaldiversityinantibodiesandT-cellreceptors.Bycontrast,homologousrecombination,althoughconceptuallymorecomplicatedthanDNAend-joining,ensuresaccuraterepairbecausetheundamagedsisterchromatidorhomologouschromosomeisusedasatemplate・WhileaccuraterepairseemscrucialforaunicellularorganismsuchasS.cerevisiae,andmammaliangermandstemcells;inaccuraterepairmightbetoleratedbydifferentiatedsomaticcellsgiventhatalargefractionoftheirgenomeisnoIongerfunctional.However,misalignmentofrepetitiveDNAsequencesmightcomplicatehomologousrecombinationinmammaliancells一asthiscouldleadtodeletionsandtranslocations.Therefore,itwouldbeadvantageoustorestricthomologousrecombinationtotheSphaseofthecellcyclesuchthatrecombinationwilloccurpreferentiallybetweenreplicatingchromatids.TheexpressionpatternoftheRAD51andRAD54genesisconsistentwiththisnotion10.Inaddition,vertebratecellsthatarehighlyactiveinhomologousrecombination,suchaschickenDT40cellsandmouseembryonicstemcells,haveveryshortG1andG2phases.InadditiontotheirroleinDSBrepair,theproteinsdescribedinthisreviewcouldalsofunctioninothercellularprocesses・Forinstance,ScKu70/80pandRad50-Mrell-Xrs2palsofunctioinintelomerelengthmaintenance52,60,61.DifferentialrolesofDSBrepairproteinsinothercellularprocessescouldhelpexplaindifferencesinphenotypesofmiceandcellsinwhichthethreegenesencodingDNA-PKhavebeenmutated.InactivationofDNA-PKCS,KU70orKU80causesSCIDandhypersensitivitytoIR.However,KU80-deficientmicedisplaygrowthretardationandarenotparticularlycancerprone,whereasDNA-PKCS-deficientmicehaveanormalgrowthratebutareverycancerprone37,39,40,62.MutationsinthegenesencodingcomponentsoftheRAD50-MRE11-NBS1\ncomplexalsoresultindistinetpheno-types.DisruptionofmouseMRE11resultsin"vi ablecells63,whereasNijmegenbreakagesyndromepatientscarryapparentnullmutationsinNBS158,59.FutureprospectsThecommonuseofIRinanti-tumourradiotherapymakesitparamounttogainfurtherinsightintothemolecularintricaciesofIR-inducedDNAdamagerepair.Inthepastfewyears,impressiveprogresshasbeenmadeinideratifyinggenesthatareimportantfortherepairofDSBs.ItisnowclearthatgenesinvolvedinthetwomajorpathwaysforDSBrepairityofthesegeneshasprovidedtheessentialtoolstotranslatetheresultsfromtheinitialgeneticanalysestomolecularmechanismsofDSBrepair.Currently,rapidprogressisbeingmadeintheanalysisofbiochemicalactivitiesofthegeneproductsinvolvedinbothDSBrepairpathways.AmajorchallengeaheadistheevaluationoftherelativecontributionofthedifferentDSBrepairpathwaystogenomicstabilityandsurvivalafterexposuretoIRindifferentmammaliantissuesandtumours.Givenpotentialdiffer-encesinfidelityofrepairthroughthedifferentpathways,thisinformationiscrucialinassessingtherisksarisingfromradiationexposure.