- 70.16 KB
- 2022-08-13 发布
- 1、本文档由用户上传,淘文库整理发布,可阅读全部内容。
- 2、本文档内容版权归属内容提供方,所产生的收益全部归内容提供方所有。如果您对本文有版权争议,请立即联系网站客服。
- 3、本文档由用户上传,本站不保证质量和数量令人满意,可能有诸多瑕疵,付费之前,请仔细阅读内容确认后进行付费下载。
- 网站客服QQ:403074932
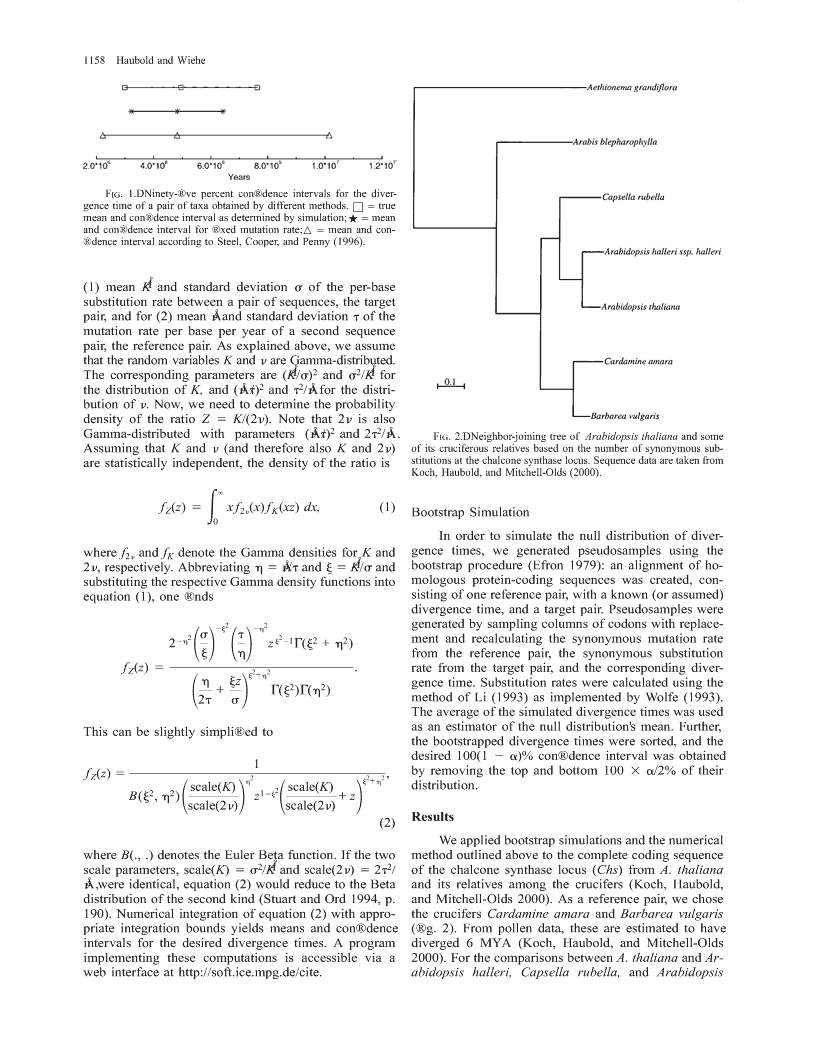
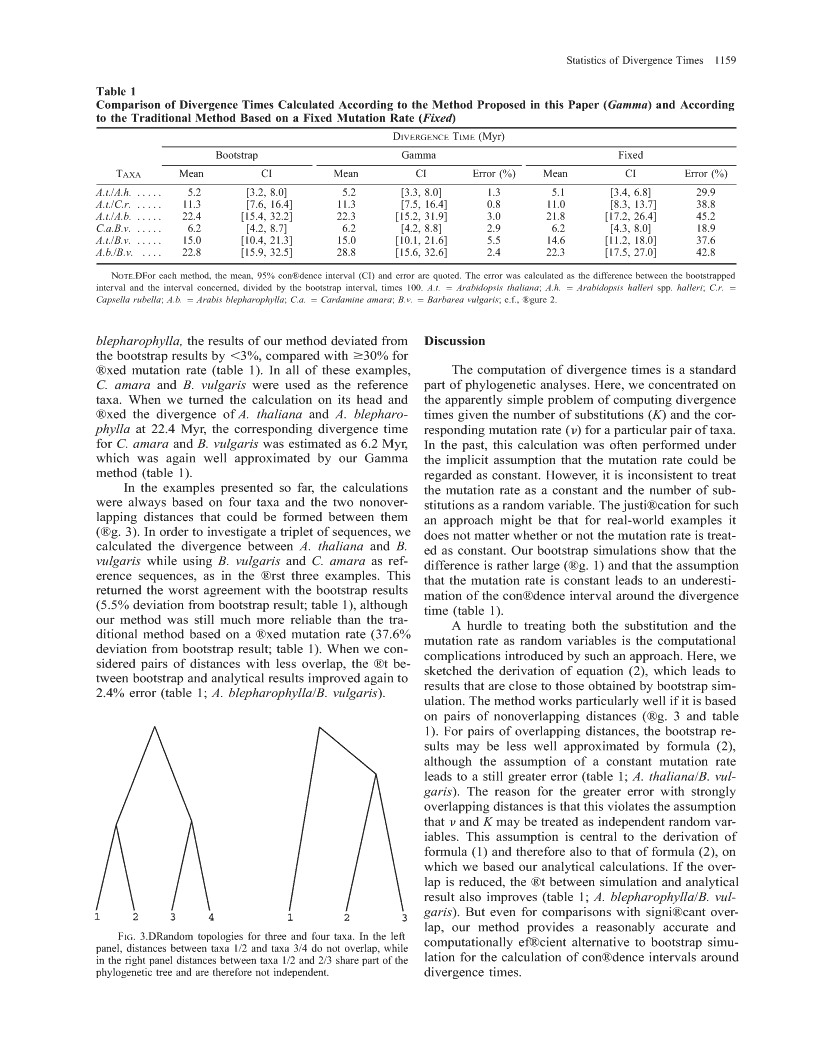
StatisticsofDivergenceTimesBernhardHauboldandThomasWieheMax-Planck-InstitutfuÈrChemischeOÈkologie,Jena,GermanyGiventhenumberofnucleotidesubstitutionsbetweentwospecies(K)andthesubstitutionraten,theexpectationofthecorrespondingdivergencetimeisusuallycalculatedasK/(2n).Thisisstrictlytrueonlyifnisregardedasaconstantbecausetheratiooftworandomvariables,suchasK/(2n),hasdistributionalpropertiesdifferentfromthoseofthedistributionofK.Therefore,boththemeanandanycon®denceintervalfordivergencetimesareunknowninthissituation.WemodelthedistributionofKandnusingtheGammadistributionandcalculatethemeanand95%con®denceintervalforthecorrespondingdivergencetime.Thesecalculationsarecomparedwithresultsob-tainedbybootstrappingsequencedatafromthemodelplantArabidopsisthalianaanditsrelatives.Weshowthatfornonoverlappingpairsofphylogeneticdistances,ourmethodapproachesthebootstrapresultsveryclosely.Incontrast,regardingthemutationrateasaconstantleadstostrongunderestimationofthecon®denceinterval.Animplementationofourmethodofcomputingdivergencetimesisaccessiblethroughawebinterfaceathttp://www.soft.ice.mpg.de/cite.IntroductionThequestionofhowdistantlytwotaxaaresepa-divergencetimeas[2.223106,1.023107].Regardingratedfromtheirlastcommonancestorisprobablyasoldthemutationrateas®xedledtotheunderestimationofasbiologyitself.WithDNAdata,theexpecteddiver-thisintervalbyapproximately30%(®g.1).gencetimebetweentaxaiandj,Tij,canbecomputedSteel,Cooper,andPenny(1996)recognizedthisinastraightforwardwayifthemutationrateisregardedproblemandsuggestedthefollowingsolution:let[n1,asconstant:n2]bea100(12a/2)%con®denceintervalforn.Inthiscase,[K1/(2n2),K2/(2n1)]isthoughttobea100(1E(Kij)2a)%con®denceintervalofthedivergencetime(Steel,E(Tij)5,Cooper,andPenny1996).Thismethoddidindeedlead23E(n)toawidercon®denceintervalthanthatobtainedfor®xedn,butthistime,theintervalwasabout70%widerwhereKijisthenumberofsubstitutionspersitebetweenthanexpected(®g.1).Furthermore,themathematicaltaxaiandj,andnisthenumberofsubstitutionspersitejusti®cationfortheirproposedmethodisunclear.Intheperyear.Ifwefurtherlet[K1,K2]bea100(12a)%following,weshallpresentasolutiontothisproblemcon®denceintervalforKij,thecorrespondingcon®denceandapplyittothedivergencebetweenthemodelplantintervalforTijisArabidopsisthalianaanditsrelativesamongthecrucifers.[T12,T]5[K1/(2n),K2/(2n)].MaterialsandMethodsDerivationofaProbabilityDensityFunctionoftheHowever,nisusuallyestimatedfromthenumberDivergenceTimeofsubstitutionsbetweenapairoftaxathatcanbedated,e.g.,byreferencetofossildata,andhenceisarandomVariationinthesubstitutionrateamongsitesalongvariableitself.ThiscomplicatesthecomputationofbothaDNAsequenceisoftenmodeledbyaGammaprob-themeanandthecon®denceintervalfordivergenceabilitydistribution(Yang1996).Inthesemodels,thetimes.Asweshallsee,thedifferencebetweenregardingnumberofsubstitutionsisnegativebinomiallydistrib-nasarandomvariableandregardingitasconstantisuted(StuartandOrd1994,p.182).Equatingthe®rstmuchmoremarkedforthecon®denceintervalthanforandsecondmomentsofthenegativebinomialdistribu-tionwiththoseofaGammadistribution,thetwoparam-themean.etersa(shape)andb(scale)oftheGammadistributionInordertodevelopanintuitionforthecalculationcanbeuniquelydetermined.Forthebiologicallyrea-ofdivergencetimes,wecarriedoutasetofexploratorysonableparameterstested,theGammadistributionpro-simulations.LetKandnbenormallydistributedrandomvariableswithmeans0.141and1.4631028andstan-videsanexcellentapproximationofthenegativebino-mialdistribution.Furthermore,ifthenumberofsubsti-darddeviations0.024and0.02531029.Thesearebi-tutionsinagene(H)isGamma-distributed,thentheologicallymeaningfulvalues.Wedrew105randomnumberofsubstitutionspersiteK5H/n,wherende-numbersfromthesedistributionsandcalculatedtheex-notesthenumberofsites,isalsoGamma-distributed.pected95%con®denceintervalforthecorrespondingOurmethodstartswiththisassumption.ThedensityfunctionoftheGammadistributionisKeywords:divergencetime,substitutionrate,Gammadistribu-tion,Arabidopsisthaliana.xb2aexp122xa21Addressforcorrespondenceandreprints:BernhardHaubold,bMax-Planck-InstitutfuÈrChemischeOÈkologie,Carl-Zeiss-Promenadef(x)5,x$0.10,D-07745Jena,Germany.E-mail:haubold@ice.mpg.de.G(a)Mol.Biol.Evol.18(7):1157±1160.2001q2001bytheSocietyforMolecularBiologyandEvolution.ISSN:0737-4038Weassumethatweareprovidedwithmeasurementsfor1157\n1158HauboldandWieheFIG.1.ÐNinety-®vepercentcon®denceintervalsforthediver-gencetimeofapairoftaxaobtainedbydifferentmethods.h5truemeanandcon®denceintervalasdeterminedbysimulation;.5meanandcon®denceintervalfor®xedmutationrate;n5meanandcon-®denceintervalaccordingtoSteel,Cooper,andPenny(1996).(1)meanKÅandstandarddeviationsoftheper-basesubstitutionratebetweenapairofsequences,thetargetpair,andfor(2)meannÅandstandarddeviationtofthemutationrateperbaseperyearofasecondsequencepair,thereferencepair.Asexplainedabove,weassumethattherandomvariablesKandnareGamma-distributed.Thecorrespondingparametersare(KÅ/s)2ands2/KÅforthedistributionofK,and(nÅ/t)and22t/nÅforthedistri-butionofn.Now,weneedtodeterminetheprobabilitydensityoftheratioZ5K/(2n).Notethat2nisalsoGamma-distributedwithparameters(nÅ/t)and222t/nÅ.FIG.2.ÐNeighbor-joiningtreeofArabidopsisthalianaandsomeAssumingthatKandn(andthereforealsoKand2n)ofitscruciferousrelativesbasedonthenumberofsynonymoussub-arestatisticallyindependent,thedensityoftheratioisstitutionsatthechalconesynthaselocus.SequencedataaretakenfromKoch,Haubold,andMitchell-Olds(2000).`fZ(z)5Exf2n(x)fK(xz)dx,(1)BootstrapSimulation0Inordertosimulatethenulldistributionofdiver-wheref2nandfKdenotetheGammadensitiesforKandgencetimes,wegeneratedpseudosamplesusingthe2n,respectively.Abbreviatingh5nÅ/tandj5KÅ/sandbootstrapprocedure(Efron1979):analignmentofho-substitutingtherespectiveGammadensityfunctionsintomologousprotein-codingsequenceswascreated,con-equation(1),one®ndssistingofonereferencepair,withaknown(orassumed)divergencetime,andatargetpair.Pseudosampleswere2j222hgeneratedbysamplingcolumnsofcodonswithreplace-22h22stzj2122G(j+h)mentandrecalculatingthesynonymousmutationrate1212jhfromthereferencepair,thesynonymoussubstitutionfZ(z)522.ratefromthetargetpair,andthecorrespondingdiver-j+hhjzgencetime.Substitutionrateswerecalculatedusingthe12+G(j22)G(h)2tsmethodofLi(1993)asimplementedbyWolfe(1993).Theaverageofthesimulateddivergencetimeswasusedasanestimatorofthenulldistribution'smean.Further,Thiscanbeslightlysimpli®edtothebootstrappeddivergencetimesweresorted,andthedesired100(12a)%con®denceintervalwasobtained1fZ(z)5222,byremovingthetopandbottom1003a/2%oftheirhj+hscale(K)2scale(K)distribution.B(j22,h)1212z12j+zscale(2n)scale(2n)Results(2)WeappliedbootstrapsimulationsandthenumericalwhereB(.,.)denotestheEulerBetafunction.Ifthetwomethodoutlinedabovetothecompletecodingsequencescaleparameters,scale(K)5s2/KÅandscale(2n)52t2/ofthechalconesynthaselocus(Chs)fromA.thaliananÅ,wereidentical,equation(2)wouldreducetotheBetaanditsrelativesamongthecrucifers(Koch,Haubold,distributionofthesecondkind(StuartandOrd1994,p.andMitchell-Olds2000).Asareferencepair,wechose190).Numericalintegrationofequation(2)withappro-thecrucifersCardamineamaraandBarbareavulgarispriateintegrationboundsyieldsmeansandcon®dence(®g.2).Frompollendata,theseareestimatedtohaveintervalsforthedesireddivergencetimes.Aprogramdiverged6MYA(Koch,Haubold,andMitchell-Oldsimplementingthesecomputationsisaccessibleviaa2000).ForthecomparisonsbetweenA.thalianaandAr-webinterfaceathttp://soft.ice.mpg.de/cite.abidopsishalleri,Capsellarubella,andArabidopsis\nStatisticsofDivergenceTimes1159Table1ComparisonofDivergenceTimesCalculatedAccordingtotheMethodProposedinthisPaper(Gamma)andAccordingtotheTraditionalMethodBasedonaFixedMutationRate(Fixed)DIVERGENCETIME(Myr)BootstrapGammaFixedTAXAMeanCIMeanCIError(%)MeanCIError(%)A.t./A.h......5.2[3.2,8.0]5.2[3.3,8.0]1.35.1[3.4,6.8]29.9A.t./C.r......11.3[7.6,16.4]11.3[7.5,16.4]0.811.0[8.3,13.7]38.8A.t./A.b......22.4[15.4,32.2]22.3[15.2,31.9]3.021.8[17.2,26.4]45.2C.a.B.v......6.2[4.2,8.7]6.2[4.2,8.8]2.96.2[4.3,8.0]18.9A.t./B.v......15.0[10.4,21.3]15.0[10.1,21.6]5.514.6[11.2,18.0]37.6A.b./B.v.....22.8[15.9,32.5]28.8[15.6,32.6]2.422.3[17.5,27.0]42.8NOTE.ÐForeachmethod,themean,95%con®denceinterval(CI)anderrorarequoted.Theerrorwascalculatedasthedifferencebetweenthebootstrappedintervalandtheintervalconcerned,dividedbythebootstrapinterval,times100.A.t.5Arabidopsisthaliana;A.h.5Arabidopsishallerispp.halleri;C.r.5Capsellarubella;A.b.5Arabisblepharophylla;C.a.5Cardamineamara;B.v.5Barbareavulgaris;c.f.,®gure2.blepharophylla,theresultsofourmethoddeviatedfromDiscussionthebootstrapresultsby,3%,comparedwith$30%for®xedmutationrate(table1).Inalloftheseexamples,ThecomputationofdivergencetimesisastandardC.amaraandB.vulgariswereusedasthereferencepartofphylogeneticanalyses.Here,weconcentratedontaxa.Whenweturnedthecalculationonitsheadandtheapparentlysimpleproblemofcomputingdivergence®xedthedivergenceofA.thalianaandA.blepharo-timesgiventhenumberofsubstitutions(K)andthecor-phyllaat22.4Myr,thecorrespondingdivergencetimerespondingmutationrate(n)foraparticularpairoftaxa.forC.amaraandB.vulgariswasestimatedas6.2Myr,Inthepast,thiscalculationwasoftenperformedunderwhichwasagainwellapproximatedbyourGammatheimplicitassumptionthatthemutationratecouldbemethod(table1).regardedasconstant.However,itisinconsistenttotreatIntheexamplespresentedsofar,thecalculationsthemutationrateasaconstantandthenumberofsub-werealwaysbasedonfourtaxaandthetwononover-stitutionsasarandomvariable.Thejusti®cationforsuchlappingdistancesthatcouldbeformedbetweenthemanapproachmightbethatforreal-worldexamplesit(®g.3).Inordertoinvestigateatripletofsequences,wedoesnotmatterwhetherornotthemutationrateistreat-calculatedthedivergencebetweenA.thalianaandB.edasconstant.OurbootstrapsimulationsshowthatthevulgariswhileusingB.vulgarisandC.amaraasref-differenceisratherlarge(®g.1)andthattheassumptionerencesequences,asinthe®rstthreeexamples.Thisthatthemutationrateisconstantleadstoanunderesti-returnedtheworstagreementwiththebootstrapresultsmationofthecon®denceintervalaroundthedivergence(5.5%deviationfrombootstrapresult;table1),althoughtime(table1).ourmethodwasstillmuchmorereliablethanthetra-Ahurdletotreatingboththesubstitutionandtheditionalmethodbasedona®xedmutationrate(37.6%mutationrateasrandomvariablesisthecomputationaldeviationfrombootstrapresult;table1).Whenwecon-complicationsintroducedbysuchanapproach.Here,wesideredpairsofdistanceswithlessoverlap,the®tbe-sketchedthederivationofequation(2),whichleadstotweenbootstrapandanalyticalresultsimprovedagaintoresultsthatareclosetothoseobtainedbybootstrapsim-2.4%error(table1;A.blepharophylla/B.vulgaris).ulation.Themethodworksparticularlywellifitisbasedonpairsofnonoverlappingdistances(®g.3andtable1).Forpairsofoverlappingdistances,thebootstrapre-sultsmaybelesswellapproximatedbyformula(2),althoughtheassumptionofaconstantmutationrateleadstoastillgreatererror(table1;A.thaliana/B.vul-garis).ThereasonforthegreatererrorwithstronglyoverlappingdistancesisthatthisviolatestheassumptionthatnandKmaybetreatedasindependentrandomvar-iables.Thisassumptioniscentraltothederivationofformula(1)andthereforealsotothatofformula(2),onwhichwebasedouranalyticalcalculations.Iftheover-lapisreduced,the®tbetweensimulationandanalyticalresultalsoimproves(table1;A.blepharophylla/B.vul-garis).Butevenforcomparisonswithsigni®cantover-lap,ourmethodprovidesareasonablyaccurateandFIG.3.ÐRandomtopologiesforthreeandfourtaxa.Intheleftcomputationallyef®cientalternativetobootstrapsimu-panel,distancesbetweentaxa1/2andtaxa3/4donotoverlap,whileintherightpaneldistancesbetweentaxa1/2and2/3sharepartofthelationforthecalculationofcon®denceintervalsaroundphylogenetictreeandarethereforenotindependent.divergencetimes.\n1160HauboldandWieheAcknowledgmentsLI,W.-H.1993.Unbiasedestimationoftheratesofsynony-mousandnonsynonymoussubstitution.Mol.Evol.36:96±99.WethanktwoanonymousreviewersforcommentsSTEEL,M.A.,A.C.COOPER,andD.PENNY.1996.Con®denceonthemanuscript.ThisworkwassupportedbytheMaxintervalsforthedivergencetimeoftwoclades.Syst.Biol.PlanckSociety.45:127±134.STUART,A.,andJ.K.ORD.1994.Kendall'sadvancedtheoryofstatistics,Vol.1.Distributiontheory.6thedition.EdwardLITERATURECITEDArnold,London.WOLFE,K.H.1993.Softwareprogramli93.UniversityofDub-EFRON,B.1979.Bootstrapmethods:anotherlookatthejack-lin,ftp://acer.gen.tcd.ie/pub/khwolfe/li93.knife.Ann.Stat.7:1±26.YANG,Z.1996.Among-siteratevariationanditsimpactonKOCH,M.A.,B.HAUBOLD,andT.MITCHELL-OLDS.2000.phylogeneticanalyses.TREE11:367±372.ComparativeevolutionaryanalysisofchalconesynthaseandalcoholdehydrogenaselociinArabidopsis,Arabis,andFUMIOTAJIMA,reviewingeditorrelatedgenera(Brassicaceae).Mol.Biol.Evol.17:1483±1498.AcceptedMarch5,2001